Molecular Diagnostics, Molecular Therapy and Experimental Transplantation
The department and research group is a diagnostics and research department with the aim of further developing molecular diagnostics, molecular therapy and experimental transplantation through patient-oriented care from "bench-to-bedside" for patient-oriented care. The department offers molecular diagnostics for all known congenital, primary immunodeficiencies, as well as molecular diagnostics for selected haematopoietic defects. Contract diagnostics for immune and haematopoietic defects are provided to patients and their family members in collaboration with clinics, physicians and geneticists. The research area focuses on the elucidation of previously uncharacterised developmental and functional disorders of the lymphatic, myeloid and erythroid series, as well as autoimmune diseases and auto-inflammations.
To date, variants have been described in more than 500 human genes that can lead to inborn errors of immunity (IEI). In addition to deficiencies of the immune system, both autoimmunity and auto-inflammation can be part of the clinical manifestations of patients. The diagnosis of these diseases includes genetic analysis, in which the variants in the genomic DNA of the patients are detected, as well as the elucidation of the pathomechanistic processes that lead to the various diseases in the presence of the individual gene variants.
These pathophysiological analyses can sometimes be very complex and can lead to the use of cell models, such as human inducible stem cells, into which the individual variants can be introduced for subsequent experimental analyses using the CRISPR/Cas9 system. Time and again, new genetic defects can be identified in patients and insights into the complex functioning of an intact human immune system can be gained from the identified disorders of immunobiological processes.
Early detection of bacteraemia, identification of the pathogen and possible accompanying pathogenicity factors and antimicrobial resistance are of great clinical importance. In the context of sepsis, for example, the survival rate drops by around 8% per hour if treatment is delayed. In the context of transfusion medicine, pathogen detection plays a role, for example, in the context of stem cell transplants or in the manufacture of blood products. Contamination of blood products, especially platelet concentrates, with bacteria, viruses and other pathogens is a serious problem that affects both the quality of the transfusion product and the safety of the recipient.
In the context of sepsis, the dilemma of delayed diagnosis of infectious diseases and sepsis in particular is reflected in the current diagnostic criteria for sepsis, which require a suspected or confirmed infection in combination with an increase of two points in the Sequential Organ Failure Assessment (SOFA) score. As the name suggests, this score increases when organ failure is already recognisable. The department is developing new methods for this problem.
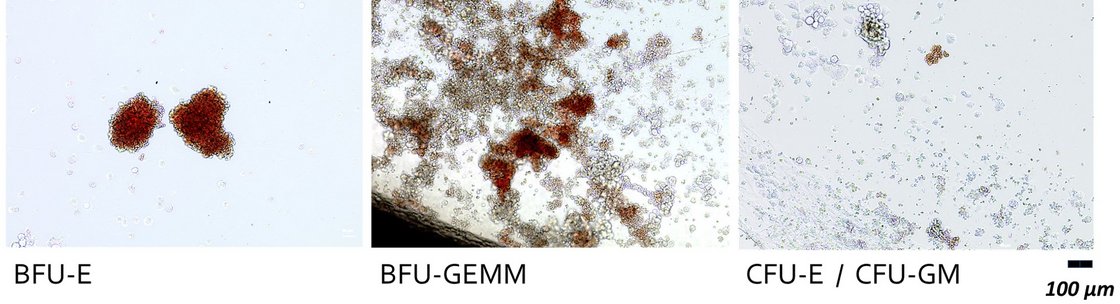
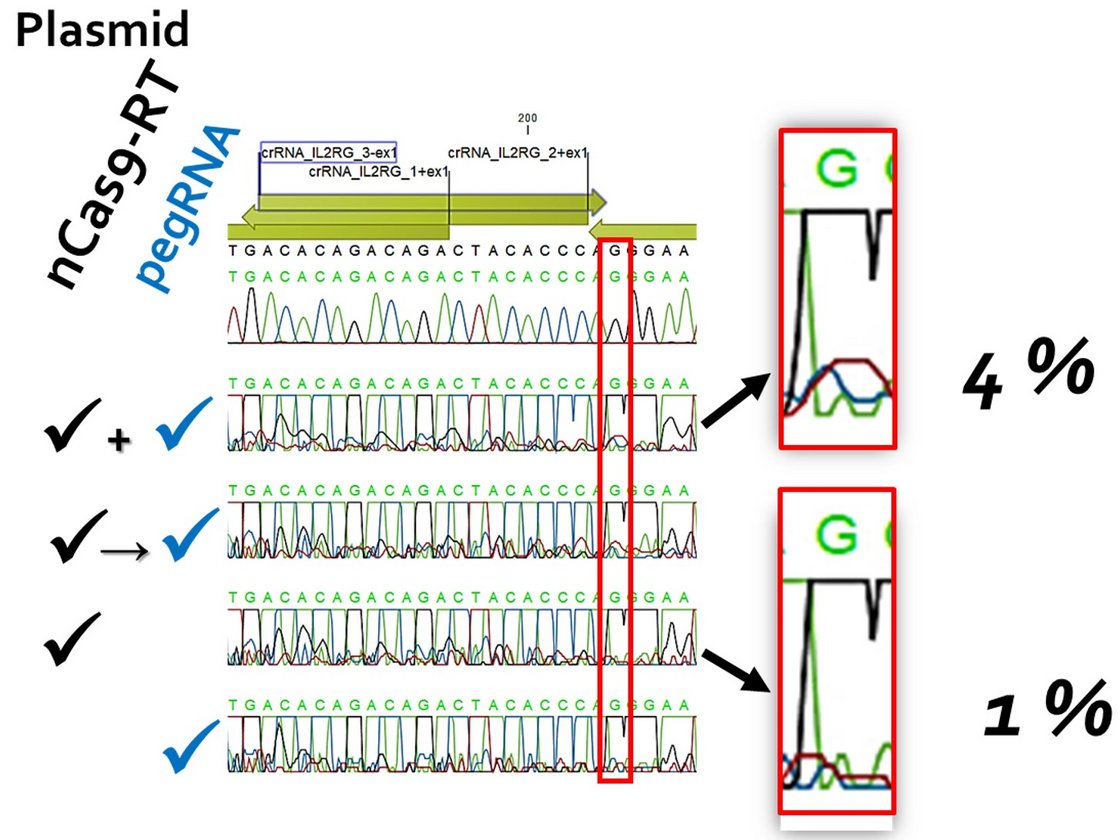
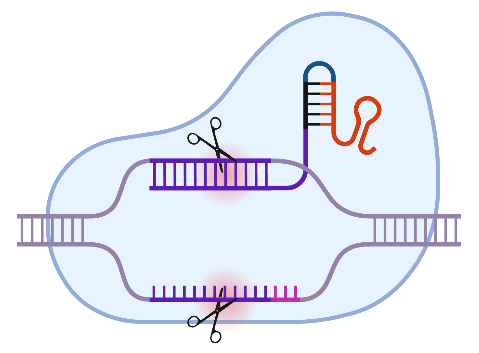
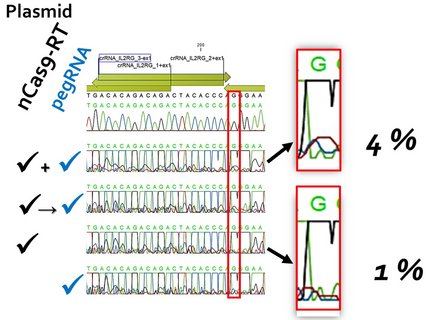
It is estimated that there are around 10,000 monogenetic human diseases. These include X-linked severe combined immunodeficiency (X-SCID), which accounts for around 50 % of all SCID cases. It is caused by mutations in the IL2RG gene. The availability of the CRISPR/Cas system has now given a significant boost to research into non-viral gene correction approaches for such diseases. Prime editing (PE) is a promising platform, not least due to the use of a Cas9 variant that introduces a single-strand break ("nick") instead of a DNA double-strand break. Further improvements to the methodology are continually being introduced. These concern both the design of the PE components and the manipulation of the target cell metabolism, for example to eliminate undesirable effects of the intrinsically necessary "mismatch" repair mechanism.
The desired goal would be to correct mutations in the IL2RG gene in human CD34+ haematopoietic stem cells ex vivo with efficiencies > 50 %. After treatment, the cells should a) retain their stem cell properties and b) their capacity for long-term repopulation. Basically, we want to define a protocol that requires as few molecular tools as possible. The first task should therefore be to improve efficiency within the given experimental framework.
Third-generation sequencing (TGS) refers to a novel sequencing technology that can sequence DNA or RNA fragments in their entire length. While TGS has disadvantages in terms of processing time and throughput compared to conventional second-generation sequencing technology, TGS offers advantages in terms of sequencing complex genetic questions, as it is both a scalable technology and enables the analysis of long DNA or RNA fragments.
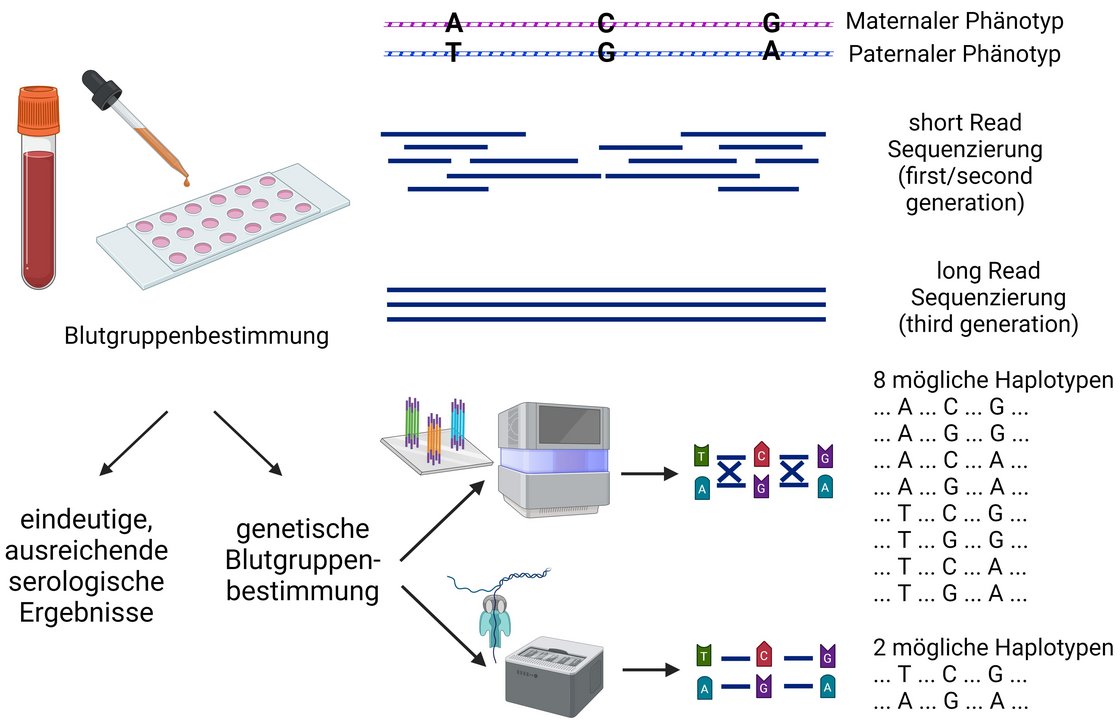
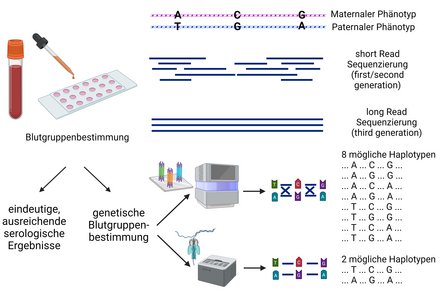
Long reads and other advantages enable a number of applications related to transfusion medicine. Current areas of application include the sequencing of microbial pathogens, including virulence and resistance determination. The technique is also used to resolve some genes such as the NCF1 gene, which is difficult to analyse using conventional short-read sequencing due to the problem of two pseudogenes. This and the resolution of haplotypes is also a field of application in blood group diagnostics, for example as shown in the Rhesus blood group system.
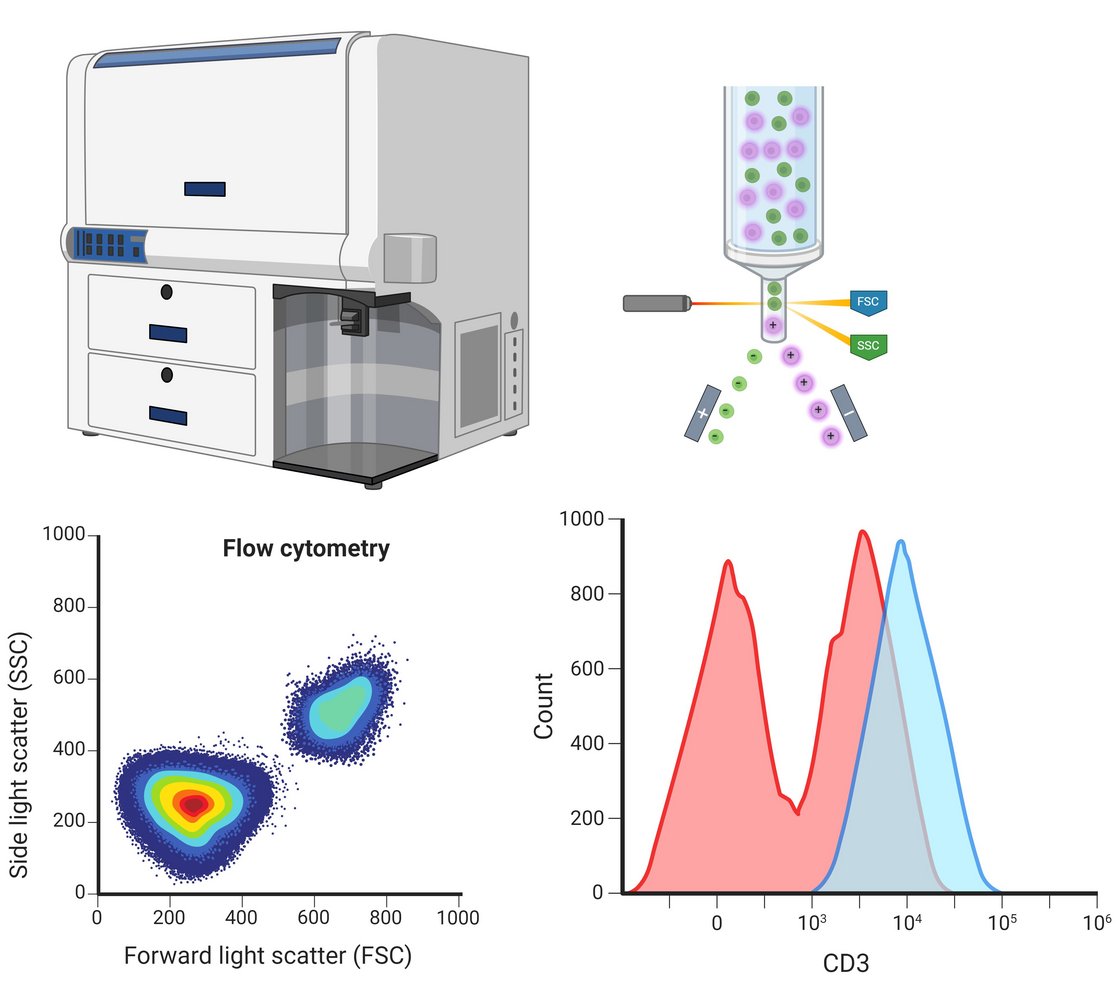
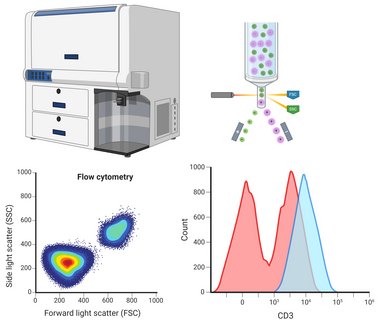
The flow cytometry method is mainly used in the department to analyse leukocytes, thrombocytes and stem cells. One specific application example is the elucidation of congenital defects of the immune system in order to quantitatively analyse haematopoietic cells using surface markers in a multi-parametric manner and to purify target cells by sorting. Four flow cytometers are available for this purpose: Two pure analysers with two lasers and five evaluation channels or with five lasers and 30 evaluation channels as well as two cell sorters with two lasers and eight evaluation channels or with three lasers and 15 evaluation channels.
Our department uses a broad repertoire of molecular genetic and molecular biological methods as part of its research and diagnostic work. A central group of molecular genetic and molecular biological methods involves working with nucleic acids and analysing them. This includes the polymerase chain reaction (PCR), which enables the selective amplification of certain segments of DNA. Numerous advanced variants of PCR, such as reverse transcriptase quantitative PCR (RT-qPCR), which enables the detection and quantification of specific RNA sequences, have been established in the department.
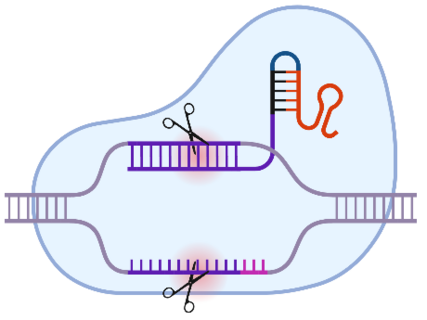
There is also a microbiological laboratory in which, for example, the recombinant expression of proteins from genetically modified E. coli bacteria is carried out. Other methods enable the expression products to be analysed: Western blotting, for example, allows the identification of the target protein in a protein mixture based on its size and specific fluorescence-coupled antibodies against the protein. These methods are frequently used in the department for the expression of recombinant Cas9 proteins. In combination with a target-specific guide molecule, the guide RNA (gRNA), targeted cuts can be made in the genome. With further modifications of the technique, even specific modifications of certain bases or the capture of targeted DNA segments from the entire genome are possible.
The department is also equipped for the development and realisation of cell-based assays. Several cell culture banks and incubators enable sterile cultivation of human cells. Various microscopes, including a laser scanning microscope, are available for follow-up analyses.
The availability of this wide range of established methods and equipment enables the department to work on a wide variety of issues, both in routine diagnostics projects and in the field of research.
Our department uses a wide range of sequencing techniques to analyse DNA and RNA molecules. In general, a well thought-out mix of sequencing technologies of each generation enables projects to be realised and optimally processed according to requirements.
Our sequencing platforms include
- First Generation Sequencing: The classical method according to Sanger, which offers high accuracy and reliability. This method is suitable for sequencing short DNA segments, such as single genes or exons. We use a modern capillary electrophoresis device that guarantees high resolution and sensitivity as well as a high sample throughput of up to 96 samples per hour.
- Second Generation Sequencing: This method with extremely high capacity is suitable for sequencing extremely large numbers of samples or large areas of the human genome thanks to massive parallelisation and a high sequencing capacity. In particular, the sequencing of large gene panels in various patient cohorts or cell populations is realised here for a wide range of research projects.
- Third Generation Sequencing: This technique enables the sequencing of individual nucleic acid molecules using very long read lengths. Various device models are available in the department, which enable a rapid change in terms of capacity, duration and cost efficiency. This technology is used for sequencing and analyses of complex or repetitive genome regions, large structural changes and transcripts.

The human immune system is a complex network of cells and organs that plays a crucial role in the defence against diseases. One of the central organs of the immune system is the thymus, which plays a key role in the formation and maturation of T cells. In recent years, research has made significant progress in the development of "artificial thymus organoids" (ATO). This innovative technology allows us to focus in detail on the disruption of T cell development in various genetic defects.
The thymus organoids are composed of a genetically optimised murine stromal cell line and human haematopoietic stem cells. These CD34-positive stem cells can either be derived from T-cell-deficient patients and control subjects or we can generate them from human induced pluripotent stem cells (hIPSC) into which genetic variants of interest are introduced using CRISPR/Cas technology.
The ATOs allow us to precisely monitor T cell differentiation across a series of defined progenitor stages. The detection and classification of these different progenitor populations is carried out in our laboratory using flow cytometry. In this way, we can very precisely assign blockages in development in the presence of the various genetic defects to individual T cell differentiation stages.
We can then isolate conspicuous cell populations using flow cytometry-based cell sorting and then comprehensively characterise them genetically (e.g. gene expression analyses using RNASeq) as well as cell and molecular biologically (e.g. determination of surface markers, activation assays, signal transduction analyses) in order to comprehensively analyse and clarify the pathomechanisms underlying the individual congenital disorders of the T-cellular immune system.
Teaching and theses
We regularly offer internship semesters, bachelor's theses, master's theses and doctoral dissertations. The theses are closely supervised by the scientists, technical staff and the head of department, who provide an insight into our wide range of methods. Due to the complexity of the topics, a longer commitment (9 months, in individual cases 3-6 months) is usually required. If you are interested, please send a cover letter, CV and previous grades to the head of department. In addition, the department is involved in teaching at the Institute of Transfusion Medicine and in the teaching research group of the Medical Faculty of the University of Ulm.
Co-operations
- BioLAGO e.V. - The health network
- Dr D. Jovanovski, Prof. Dr A. Liebold (Department of Cardiothoracic and Vascular Surgery, University Medical Center Ulm)
- Prof Dr B. Nilsson (Department of Immunology, Genetics and Pathology, University Uppsala, Sweden)
- Prof. Dr K. Nilsson Ekdahl (Department of Immunology, Genetics and Pathology, University Uppsala, Linnæus Center of Biomaterials Chemistry, Sweden)
- Prof Dr C. Schmidt (Institute of Pharmacology of Natural Products and Clinical Pharmacology, Ulm University)
- Prof. Dr M. Weiß, Prof. Dr E. Barth (Department of Anaesthesiology and Intensive Care Medicine, University Medical Center Ulm)
- Prof. Dr. S. Ehl (Centre for Chronic Immunodeficiency, University Medical Center Freiburg)
- L. Wohlgemuth, Prof. Dr Markus Huber-Lang (Institute for Trauma Immunology, University Medical Center Ulm)
Research funding
- Else Kröner-Fresenius Foundation
- DRK Blood Donation Service Baden-Württemberg - Hesse
Aktuelle Kontaktdaten finden Sie hier.
Abteilungsleitung:
- PD Dr. med. David Messerer, MME, MHBA (Abteilungsleitung)
- Karen Bosch, Regina Geyer (Sekretariat)
Akademische Mitarbeiterinnen und Mitarbeiter:
- Timo Dinse
- PD Dr. biol. hum. Karl Föhr
- Dr. rer. nat. Marita Führer
- Dominik Nolde
- Dr. rer. nat. Frank Radecke
- Dr. rer. nat. Sina Rometsch
- Dr. biol. hum. Rebekka Waldmann
Laborantinnen und Laboranten bzw. technische Angestellte:
- Katja Bahle
- Tatjana Bischoff
- Sylvia Braun
- Philipp Hoffelner
- Gabriele Keller
- Simone Mosch
- Ingrid Peter
- Melanie Riecker
- Martina Schlaier
Promotionsstudierende
- Urs Bayer (Dr. med.)
- Vincent Kramer (Dr. rer. nat.)
- Nils Oleszewski (Dr. med.)
- Alexander Timm (Dr. med.)
- Emmanuel Treiber (Dr. med.)
Gastärzte
- Dr. med. Christian Braun